Using odds scaling to accomodate bounded SSDs via ssdtools
Joe Thorley, Rebecca Fisher and David Fox
2025-06-13
Source:vignettes/bounded.Rmd
bounded.RmdIntroduction
Currently ssdtools and the associated shiny
only includes distributions that are positive (do not include 0) but can
exceed 1. This is appropriate for most concentration data that are
typically used to derive GVs. However, in the case of direct toxicity
assessment of effluents with low toxicity which are inherently bounded
between 0 and 1 (depending on whether they represent 0% or 100% sample)
a different approach is required, using appropriately bounded
statistical distribution(s) for the SSD.
We hypothesize that while the impact of using SSDs unbounded to the right may be small for effluents with a high level of toxicity, this will be very high for effluents with low toxicity.
While it may be possible to add candidate bounded distribution(s) to
(shiny)ssdtools, such as the widely used
Beta() distribution, this would involve substantial coding
effort.
An alternative would be to implement an appropriate transformation of
the existing default distribution set to accommodate bounded data. One
such appropriate transformation would be to use odds
scaling. Transformation to the odds scale can be achieved using the
following function:
odds
#> function (x)
#> {
#> x/(1 - x)
#> }With back-transformation achieved via the corresponding inverse function:
inv_odds
#> function (x)
#> {
#> x/(x + 1)
#> }Here we compare undertaking an SSD based risk assessment using the
toxicity data on their original scale, and ignoring the potential issue
that these cannot theoretically take values of greater than 1, whilst
all the distributions currently in ssdtools assume the true
underlying maximum is Inf.
Methods
The R code that performs the analysis is as follows.
First, we test that our transformation and inversion functions work as expected:
x <- c(0.01, 0.2, 0.5, 0.8)
testthat::expect_identical(x, inv_odds(odds(x)))Next, we explore some case studies based on toxicity assessments of an effluent sample over time, expressed as a proportion of the tested sample.
Caste study 1
These data are based on a real world discharge example, however they are confidential, so we cannot share the context under which they were collected. However, for this particular case the values were all very low - in some cases much less than 1 percent sample (<0.01 when expressed as a proportion). As they are, these data can be used to asses the impact of accounting for bounding in the SSD in the case where input toxicity data are from the very left hand tail of their theoretical distribution and are therefore highly toxic.
| Spp | sp 1 | sp 2 | sp 3 | sp 4 | sp 5 | sp 6 | sp 7 | sp 8 | sp 9 | sp 10 | sp 11 |
| A | 0.00490 | 0.00450 | 0.00064 | 0.00330 | 0.00140 | 0.00051 | 0.00062 | 0.00260 | 0.00039 | 0.00520 | 0.01100 |
| B | 0.05500 | 0.04500 | 0.01100 | 0.01700 | 0.00940 | 0.00041 | 0.01500 | 0.00840 | 0.01500 | 0.05000 | 0.25000 |
| C | 0.02600 | 0.03000 | 0.01300 | 0.15000 | 0.06200 | 0.00035 | 0.00980 | 0.02200 | 0.11000 | 0.15000 | 0.07900 |
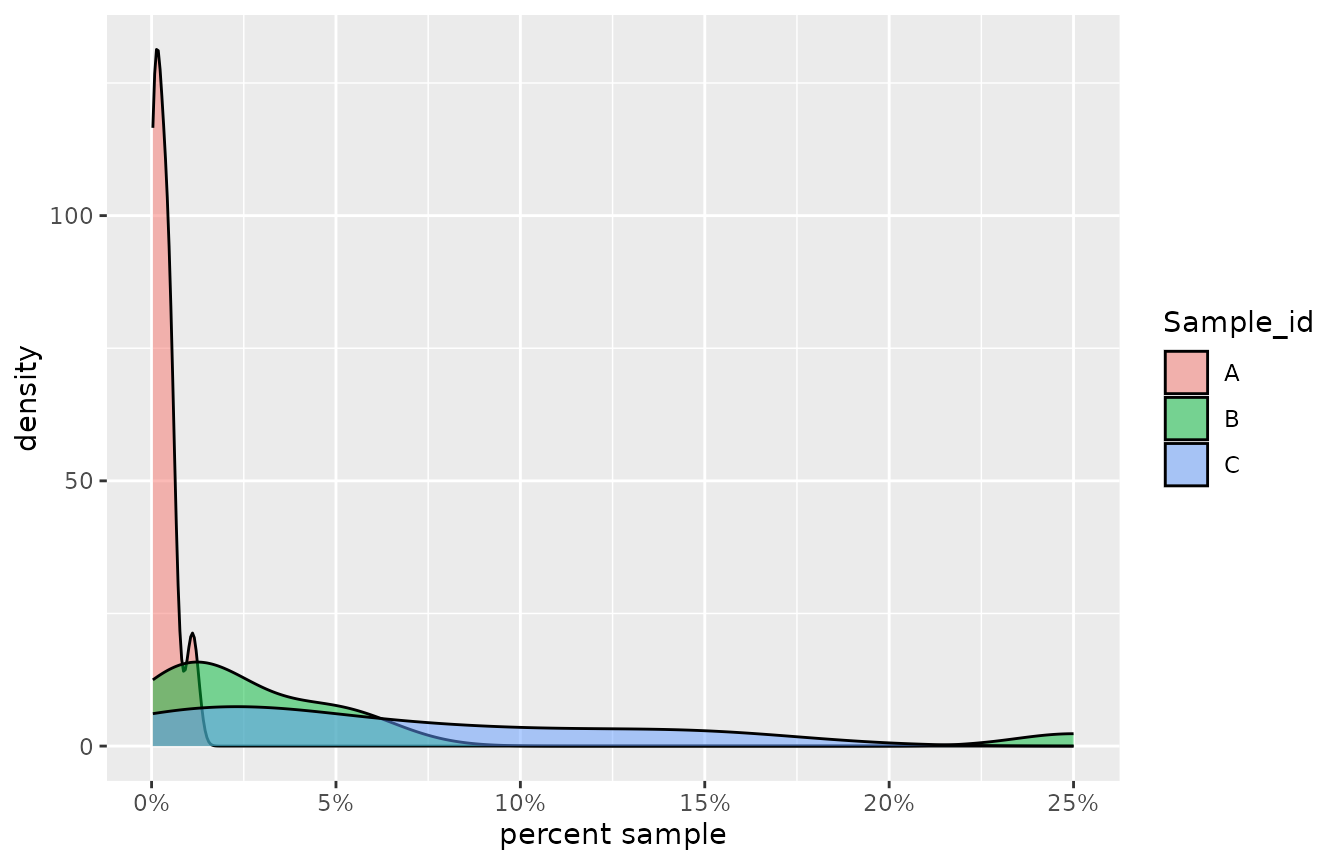
Density plots of the raw toxicity data for three highly toxic effluent samples.
Case study 2
Toxicity estimates from effluent samples may also be on the upper tail of the possible theoretical values of between 0 and 100%. Real world examples might include testing of brine effluent, where some species are relatively insensitive to the elevated salinity and may return toxicity estimates at, or near 100%. To mimic such a scenario, here we simply take the complement to the lower tail toxicity data presented previously, by simply subtracting these observed probabilities from one.
| Spp | sp 1 | sp 2 | sp 3 | sp 4 | sp 5 | sp 6 | sp 7 | sp 8 | sp 9 | sp 10 | sp 11 |
| A | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 |
| B | 0.95 | 0.96 | 0.99 | 0.98 | 0.99 | 1.00 | 0.98 | 0.99 | 0.98 | 0.95 | 0.75 |
| C | 0.97 | 0.97 | 0.99 | 0.85 | 0.94 | 1.00 | 0.99 | 0.98 | 0.89 | 0.85 | 0.92 |
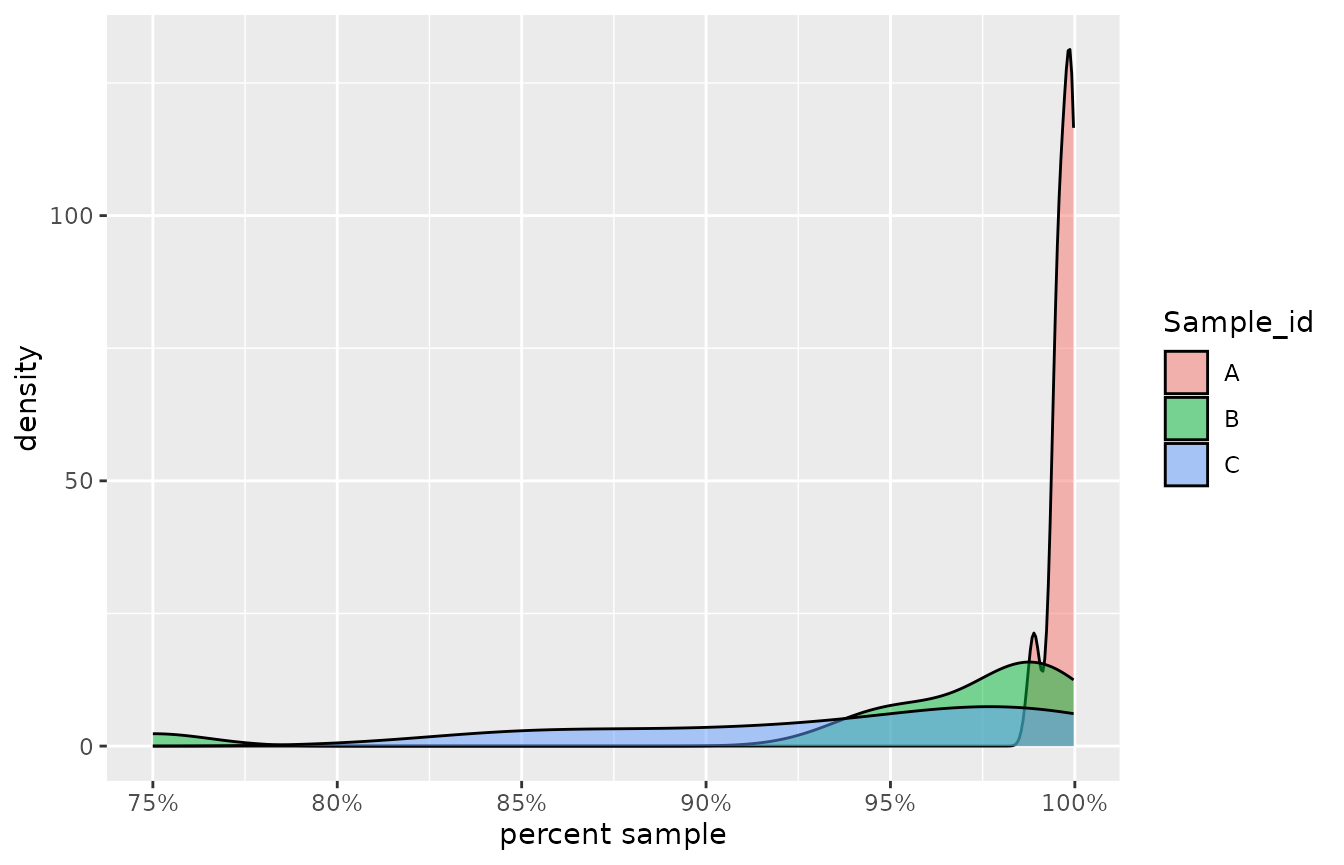
Density plots of the raw toxicity data for three low toxic effluent samples.
We used these three sets of low toxicity and high toxicity examples
to examine how the estimated number of dilutions required to reach the
different species protection values changed if we model these data on
the odds scale to account for the fact that the samples
come from a theoretically bounded distribution, compared to if we used
the original as though they are from unbounded distributions.
We fitted SSDs to both the original and odds transformed
data, using the function ssd_fit_bcanz() from the
ssdtools package in R, following the methods
outlined in the updated Australian and New Zealand Guidelines for Marine
and Freshwater Quality. The only change we made from the recommended
defaults settings was to exclude the lognormal-lognormal mixture
distribution, as this bi-modal distribution failed to fit to some of our
example data.
We obtained 80, 90, 95 and 99 species protection values (0.01, 0.05,
0.1 and 0.2 probability quantiles) from the fitted SSDs for both the
odds transformed and original fits. Based on the resulting
estimated percentages of the effluent sample, we calculated the number
of dilutions required to achieve each protection target.
Findings
We found that there was a substantial difference in the number of
dilutions required to meet the desired protection level for the 99 and
95% species protection targets, depending on if the data were modeled on
their original scaling which assumes they are unbounded, compared to
using the odds scaling.
For both high and low toxicity data examples, performing the SSD
modelling on the odds scaling resulted in a higher number
of required dilutions compared to when the data are assumed to come from
an unbounded distribution.
| Sample_id | proportion | toxicity | original | odds |
|---|---|---|---|---|
| A | 0.01 | high toxicity | 12400.334615 | 12462.907418 |
| A | 0.05 | high toxicity | 3602.662073 | 3608.552642 |
| A | 0.10 | high toxicity | 2240.143162 | 2242.273474 |
| A | 0.20 | high toxicity | 1323.234195 | 1323.714740 |
| B | 0.01 | high toxicity | 5581.959795 | 5955.621547 |
| B | 0.05 | high toxicity | 883.280182 | 913.213828 |
| B | 0.10 | high toxicity | 415.402067 | 428.420784 |
| B | 0.20 | high toxicity | 186.197147 | 190.647411 |
| C | 0.01 | high toxicity | 3185.693513 | 3743.515970 |
| C | 0.05 | high toxicity | 508.724333 | 544.892565 |
| C | 0.10 | high toxicity | 227.998540 | 235.803906 |
| C | 0.20 | high toxicity | 98.370982 | 98.440106 |
| A | 0.01 | low toxicity | 1.011012 | 1.044030 |
| A | 0.05 | low toxicity | 1.008014 | 1.012973 |
| A | 0.10 | low toxicity | 1.006644 | 1.008188 |
| A | 0.20 | low toxicity | 1.005150 | 1.004913 |
| B | 0.01 | low toxicity | 1.186611 | 1.835735 |
| B | 0.05 | low toxicity | 1.125466 | 1.229141 |
| B | 0.10 | low toxicity | 1.099455 | 1.131518 |
| B | 0.20 | low toxicity | 1.072930 | 1.068471 |
| C | 0.01 | low toxicity | 1.234257 | 1.679255 |
| C | 0.05 | low toxicity | 1.169067 | 1.261149 |
| C | 0.10 | low toxicity | 1.140361 | 1.177828 |
| C | 0.20 | low toxicity | 1.108683 | 1.110628 |
Expressed as a proportion, the bias in the number of additional
required dilutions tended to be higher for the low toxicity data (at the
upper extreme of the distribution), compared to the low toxicity data.
This would be expected given that these are the toxicity data that are
nearest to the upper bound of the distribution. However, because these
represent extremely low toxicity examples, the realised number of
required dilutions remains very low. For example, based on assuming no
bounding, 1.2 dilutions would be required to meet the 99% protection
target for the low toxicity example represented by Sample B, compared to
1.8 dilutions when modeled on the odds scale.
While the proportions of required additional dilutions are lower for
the high toxicity scenarios relative to the low toxicity scenarios, the
actual number of additional dilutions required can be very large. For
example, 3,186 dilutions would be required to meet the 99% protection
target for the high toxicity example for Sample C. In contrast 3,743
dilutions would be required when modeled on the odds
scale.
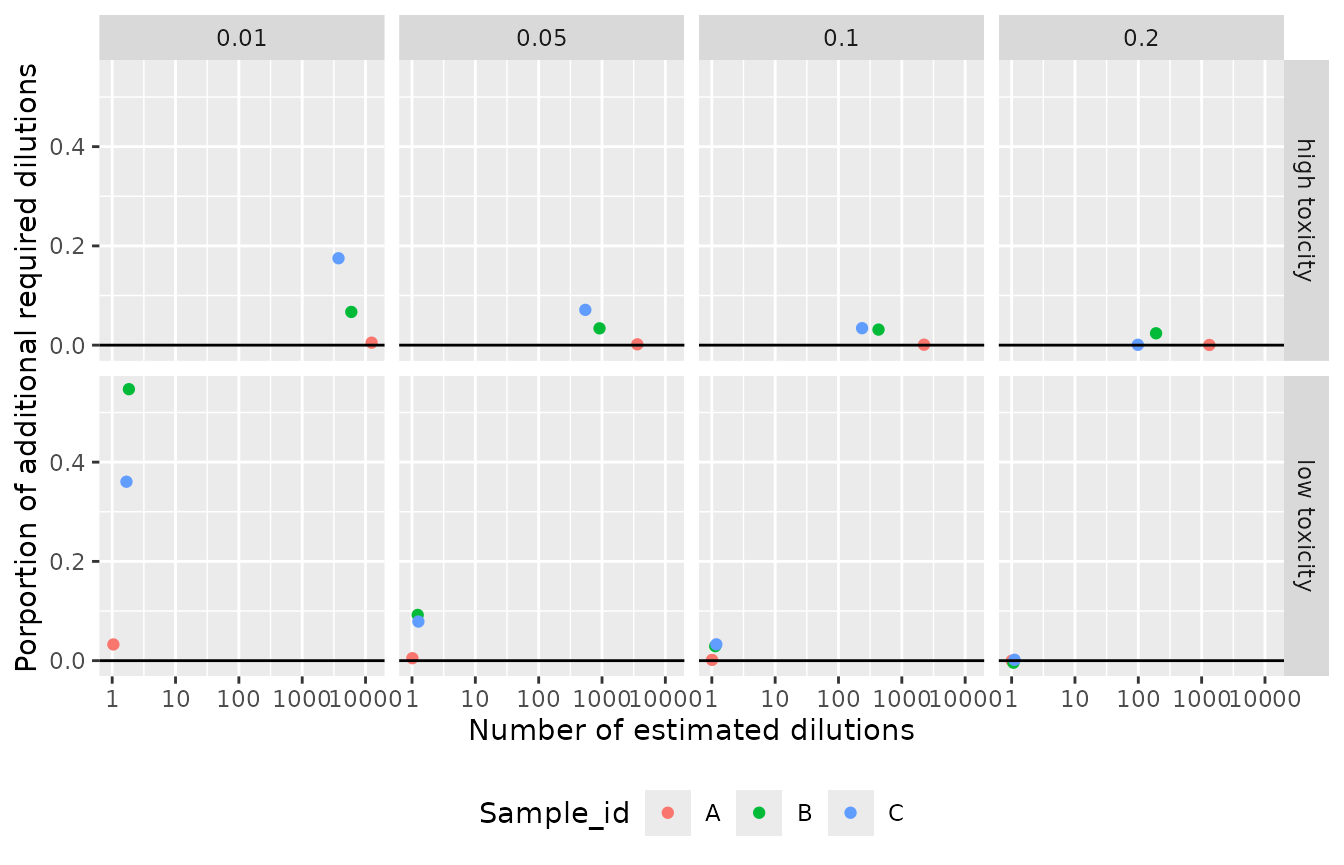
Proportion of additional required dilutions based on SSDs fitted using the original data, when compared to SSDs fitted via odds scaling.
Conclusions and Recommendations
Accommodating bounding of the SSD using odds scaling
changes the required number of dilutions to meet species protection
targets, particularly when targeting relatively high protection values
(e.g. 95 and 99% species protection). In particular, it appears that the
target number of dilutions required to meet the protection values are
underestimated when bounding is ignored, suggesting that current
practice of ignoring the fact that toxicity values come from a bounded
distribution in the case of whole effluent toxicity assessment may be
resulting in the use of guideline values that do not meet the stated
protection targets.
Session Info
The results were generated with the following packages.
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.5.0 (2025-04-11)
#> os Ubuntu 24.04.2 LTS
#> system x86_64, linux-gnu
#> ui X11
#> language en-US
#> collate C.UTF-8
#> ctype C.UTF-8
#> tz UTC
#> date 2025-06-13
#> pandoc 3.1.11 @ /opt/hostedtoolcache/pandoc/3.1.11/x64/ (via rmarkdown)
#> quarto NA
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> abind 1.4-8 2024-09-12 [1] RSPM
#> brio 1.1.5 2024-04-24 [1] RSPM
#> bslib 0.9.0 2025-01-30 [1] RSPM
#> cachem 1.1.0 2024-05-16 [1] RSPM
#> chk 0.10.0 2025-01-24 [1] RSPM
#> cli 3.6.5 2025-04-23 [1] RSPM
#> codetools 0.2-20 2024-03-31 [3] CRAN (R 4.5.0)
#> desc 1.4.3 2023-12-10 [1] RSPM
#> digest 0.6.37 2024-08-19 [1] RSPM
#> dplyr 1.1.4 2023-11-17 [1] RSPM
#> evaluate 1.0.3 2025-01-10 [1] RSPM
#> farver 2.1.2 2024-05-13 [1] RSPM
#> fastmap 1.2.0 2024-05-15 [1] RSPM
#> fs 1.6.6 2025-04-12 [1] RSPM
#> furrr 0.3.1 2022-08-15 [1] RSPM
#> future 1.58.0 2025-06-05 [1] RSPM
#> generics 0.1.4 2025-05-09 [1] RSPM
#> ggplot2 * 3.5.2 2025-04-09 [1] RSPM
#> globals 0.18.0 2025-05-08 [1] RSPM
#> glue 1.8.0 2024-09-30 [1] RSPM
#> goftest 1.2-3 2021-10-07 [1] RSPM
#> gtable 0.3.6 2024-10-25 [1] RSPM
#> htmltools 0.5.8.1 2024-04-04 [1] RSPM
#> jquerylib 0.1.4 2021-04-26 [1] RSPM
#> jsonlite 2.0.0 2025-03-27 [1] RSPM
#> knitr 1.50 2025-03-16 [1] RSPM
#> labeling 0.4.3 2023-08-29 [1] RSPM
#> lattice 0.22-6 2024-03-20 [3] CRAN (R 4.5.0)
#> lifecycle 1.0.4 2023-11-07 [1] RSPM
#> listenv 0.9.1 2024-01-29 [1] RSPM
#> magrittr 2.0.3 2022-03-30 [1] RSPM
#> Matrix 1.7-3 2025-03-11 [3] CRAN (R 4.5.0)
#> parallelly 1.45.0 2025-06-02 [1] RSPM
#> pillar 1.10.2 2025-04-05 [1] RSPM
#> pkgconfig 2.0.3 2019-09-22 [1] RSPM
#> pkgdown 2.1.3 2025-05-25 [1] any (@2.1.3)
#> pkgload 1.4.0 2024-06-28 [1] RSPM
#> plyr 1.8.9 2023-10-02 [1] RSPM
#> purrr 1.0.4 2025-02-05 [1] RSPM
#> R6 2.6.1 2025-02-15 [1] RSPM
#> ragg 1.4.0 2025-04-10 [1] RSPM
#> rbibutils 2.3 2024-10-04 [1] RSPM
#> RColorBrewer 1.1-3 2022-04-03 [1] RSPM
#> Rcpp 1.0.14 2025-01-12 [1] RSPM
#> Rdpack 2.6.4 2025-04-09 [1] RSPM
#> rlang 1.1.6 2025-04-11 [1] RSPM
#> rmarkdown 2.29 2024-11-04 [1] RSPM
#> rprojroot 2.0.4 2023-11-05 [1] RSPM
#> sass 0.4.10 2025-04-11 [1] RSPM
#> scales 1.4.0 2025-04-24 [1] RSPM
#> sessioninfo 1.2.3 2025-02-05 [1] RSPM
#> ssddata 1.0.0 2021-11-05 [1] RSPM
#> ssdtools * 2.3.0.9004 2025-06-13 [1] Github (poissonconsulting/ssdtools@17874d3)
#> stringi 1.8.7 2025-03-27 [1] RSPM
#> stringr 1.5.1 2023-11-14 [1] RSPM
#> systemfonts 1.2.3 2025-04-30 [1] RSPM
#> testthat 3.2.3 2025-01-13 [1] RSPM
#> textshaping 1.0.1 2025-05-01 [1] RSPM
#> tibble 3.3.0 2025-06-08 [1] RSPM
#> tidyr 1.3.1 2024-01-24 [1] RSPM
#> tidyselect 1.2.1 2024-03-11 [1] RSPM
#> TMB 1.9.17 2025-03-10 [1] RSPM
#> universals 0.0.5 2022-09-22 [1] RSPM
#> vctrs 0.6.5 2023-12-01 [1] RSPM
#> waldo 0.6.1 2024-11-07 [1] RSPM
#> withr 3.0.2 2024-10-28 [1] RSPM
#> xfun 0.52 2025-04-02 [1] RSPM
#> yaml 2.3.10 2024-07-26 [1] RSPM
#>
#> [1] /home/runner/work/_temp/Library
#> [2] /opt/R/4.5.0/lib/R/site-library
#> [3] /opt/R/4.5.0/lib/R/library
#> * ── Packages attached to the search path.
#>
#> ──────────────────────────────────────────────────────────────────────────────